E. coli thrives in our guts, sometimes to unfortunate effect, and it facilitates scientific advances—in DNA, biofuels, and Pfizer’s covid vaccine, to name but a few. Now this multitalented bacterium has a new trick: it can solve a classic computational maze problem using distributed computing—dividing up the necessary calculations among different types of genetically engineered cells.
This neat feat is a credit to synthetic biology, which aims to rig up biological circuitry much like electronic circuitry and to program cells as easily as computers.
The maze experiment is part of what some researchers consider a promising direction in the field: rather than engineering a single type of cell to do all the work, they design multiple types of cells, each with different functions, to get the job done. Working in concert, these engineered microbes might be able to “compute” and solve problems more like multicellular networks in the wild.
So far, for better or worse, fully harnessing biology’s design power has eluded, and frustrated, synthetic biologists. “Nature can do this (think about a brain), but we don’t yet know how to design at that overwhelming level of complexity using biology,” says Pamela Silver, a synthetic biologist at Harvard.
The study with E. coli as maze solvers, led by biophysicist Sangram Bagh at the Saha Institute of Nuclear Physics in Kolkata, is a simple and fun “toy” problem. But it also serves as a proof of principle for distributed computing among cells, demonstrating how more complex and practical computational problems might be solved in a similar way. If this approach works at larger scales, it could unlock applications pertaining to everything from pharmaceuticals to agriculture to space travel.
“As we move into solving more complex problems with engineered biological systems, spreading out the load like this is going to be an important capacity to establish,” says David McMillen, a bioengineer at the University of Toronto.
How to build a bacterial maze
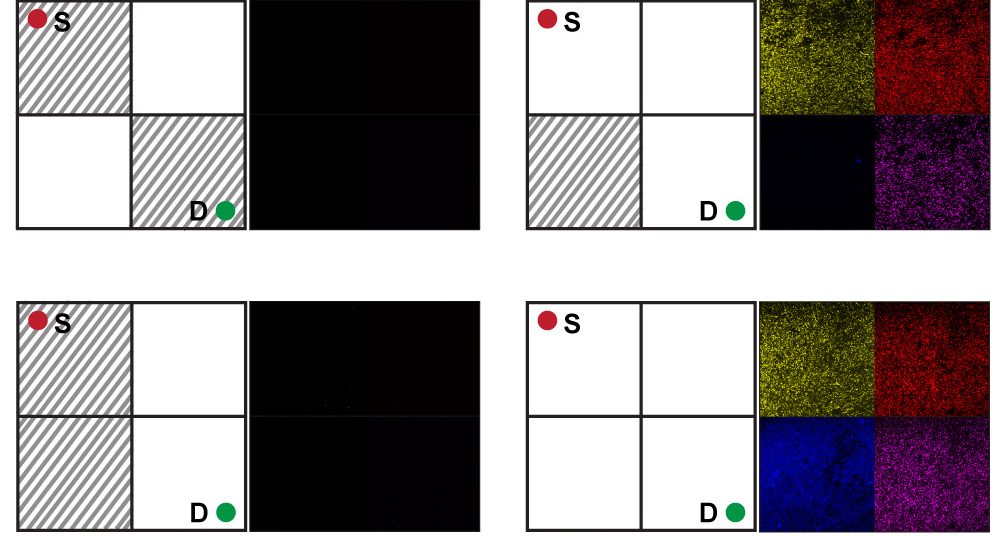
Getting E. coli to solve the maze problem involved some ingenuity. The bacteria didn’t wander through a palace labyrinth of well-pruned hedges. Rather, the bacteria analyzed various maze configurations. The setup: one maze per test tube, with each maze generated by a different chemical concoction.
The chemical recipes were abstracted from a 2 × 2 grid representing the maze problem. The grid’s top left square is the start of the maze, and the bottom right square is the destination. Each square on the grid can be either an open path or blocked, yielding 16 possible mazes.
Bagh and his colleagues mathematically translated this problem into a truth table composed of 1s and 0s, showing all possible maze configurations. Then they mapped those configurations onto 16 different concoctions of four chemicals. The presence or absence of each chemical corresponds to whether a particular square is open or blocked in the maze.
The team engineered multiple sets of E. coli with different genetic circuits that detected and analyzed those chemicals. Together, the mixed population of bacteria functions as a distributed computer; each of the various sets of cells perform part of the computation, processing the chemical information and solving the maze.
Running the experiment, the researchers first put the E. coli in 16 test tubes, added a different chemical-maze concoction in each, and left the bacteria to grow. After 48 hours, if the E. coli detected no clear path through the maze—that is, if the requisite chemicals were absent—then the system remained dark. If the correct chemical combination was present, corresponding circuits turned “on” and the bacteria collectively expressed fluorescent proteins, in yellow, red, blue, or pink, to indicate solutions. “If there is a path, a solution, the bacteria glow,” says Bagh.
KATHAKALI SARKAR AND SANGRAM BAGH
What Bagh found particularly exciting was that in churning through all 16 mazes, the E. coli provided physical proof that only three were solvable. “Calculating this with a mathematical equation is not straightforward,” he says. “With this experiment, you can visualize it very simply.”
Lofty goals
Bagh envisions such a biological computer helping in cryptography or steganography (the art and science of hiding information), which use mazes to encrypt and conceal data, respectively. But the implications extend beyond those applications to synthetic biology’s loftier ambitions.
The idea of synthetic biology dates to the 1960s, but the field emerged concretely in 2000 with the creation of synthetic biological circuits (specifically, a toggle switch and an oscillator) that made it increasingly possible to program cells to produce desired compounds or react intelligently within their environments.






Recent Comments